MATH 376 -- Probability and Statistics 2
Multiple Regression and Hypothesis testing
April 26, 2010
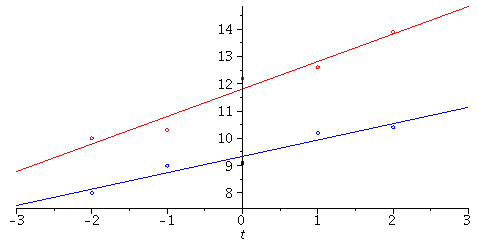
We have two strains of bacteria and want to compare their growth rates. If we do
simple regression with each data set as follows:
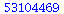 |
(1) |
![[-2, -1, 0, 1, 2]](images/HypTestMultReg_2.gif) |
(2) |
| > |
A:=[8.0,9.0,9.1,10.2,10.4]; |
![[8.0, 9.0, 9.1, 10.2, 10.4]](images/HypTestMultReg_3.gif) |
(3) |
| > |
B:=[10.0,10.3,12.2,12.6,13.9]; |
![[10.0, 10.3, 12.2, 12.6, 13.9]](images/HypTestMultReg_4.gif) |
(4) |
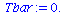 |
(5) |
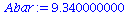 |
(6) |
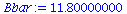 |
(7) |
| > |
SXYA:=add((times[i]-Tbar)*(A[i]-Abar),i=1..5); |
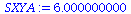 |
(8) |
| > |
SXXA:=add((times[i]-Tbar)^2,i=1..5); |
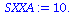 |
(9) |
| > |
Ahatbeta[1]:=SXYA/SXXA; |
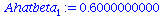 |
(10) |
| > |
Ahatbeta[0]:=Abar-Ahatbeta[1]*Tbar; |
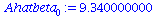 |
(11) |
| > |
SXYB:=add((times[i]-Tbar)*(B[i]-Bbar),i=1..5); |
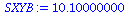 |
(12) |
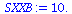 |
(13) |
| > |
Bhatbeta[1]:=SXYB/SXXB; |
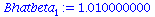 |
(14) |
| > |
Bhatbeta[0]:=Bbar-Bhatbeta[1]*Tbar; |
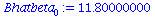 |
(15) |
| > |
APoints:=[seq([times[i],A[i]],i=1..5)]; |
![[[-2, 8.0], [-1, 9.0], [0, 9.1], [1, 10.2], [2, 10.4]]](images/HypTestMultReg_16.gif) |
(16) |
| > |
BPoints:=[seq([times[i],B[i]],i=1..5)]; |
![[[-2, 10.0], [-1, 10.3], [0, 12.2], [1, 12.6], [2, 13.9]]](images/HypTestMultReg_17.gif) |
(17) |
| > |
AP:=plot(APoints,style=point,symbol=circle,color=blue): |
| > |
BP:=plot(BPoints,style=point,symbol=circle,color=red): |
| > |
AL:=plot(Ahatbeta[0]+Ahatbeta[1]*t,t=-3..3,color=blue): |
| > |
BL:=plot(Bhatbeta[0]+Bhatbeta[1]*t,t=-3..3,color=red): |
Note: From this plot, it seems that the lines have different slopes, indicating different growth rates.
How can we test whether this is statistically significant though? Here is one method, where we
incorporate both data sets into a single set, but include a "category variable" indicating
which data set the point came from. Let ![x[1] = 0](images/HypTestMultReg_19.gif) if the measurement is from type A, and
if the measurement is from type A, and
![x[1] = 1](images/HypTestMultReg_20.gif) if the measurement is from type B. Then
if the measurement is from type B. Then ![x[2]](images/HypTestMultReg_21.gif) will indicate the time, and Y will
will indicate the time, and Y will
be the number of bacteria. The following type of linear model will
allow us to fit the straight lines for both data types, and compare their slopes:
Y = ![`+`(beta[0], Typesetting:-delayDotProduct(beta[1], x[1], true), Typesetting:-delayDotProduct(beta[2], x[2], true), `*`(Typesetting:-delayDotProduct(beta[3], x[1], true), `*`(x[2])))](images/HypTestMultReg_22.gif) +
+ 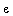
Why do we do it this way? Note: if ![x[1] = 0](images/HypTestMultReg_24.gif) (type A), we get
(type A), we get ![Y = `+`(beta[0], Typesetting:-delayDotProduct(beta[2], x[2], true))](images/HypTestMultReg_25.gif)
On the other hand if ![x[1] = 1](images/HypTestMultReg_26.gif) (type B), then we get
(type B), then we get ![Y = `+`(beta[0], beta[1], `*`(`+`(beta[2], beta[3]), `*`(x[2])))](images/HypTestMultReg_27.gif)
To compare the two slopes we just want to look at ![`+`(`+`(beta[2], beta[3]), `+`(`-`(beta[2])))](images/HypTestMultReg_28.gif) =
= ![beta[3]](images/HypTestMultReg_29.gif) . We know
. We know
how to set up a test of ![H[0]](images/HypTestMultReg_30.gif) :
: ![beta[3] = 0](images/HypTestMultReg_31.gif) versus
versus ![H[a]](images/HypTestMultReg_32.gif) :
: ![`<>`(beta[3], 0)](images/HypTestMultReg_33.gif) based on our general
based on our general
discussions(!) Now we go to the matrix formulation for the multiple regression
model. We will set up X putting the data points for type A first, then type B,
but any order would give equivalent results:
| > |
X:=Matrix([[1,0,-2,0],[1,0,-1,0],[1,0,0,0],[1,0,1,0],[1,0,2,0],[1,1,-2,-2],[1,1,-1,-1],[1,1,0,0],[1,1,1,1],[1,1,2,2]]): |
| > |
Y:=Matrix([[8.0],[9.0],[9.1],[10.2],[10.4],[10.0],[10.3],[12.2],[12.6],[13.9]]): |
The normal equations:
| > |
XtX:=Multiply(Transpose(X),X); |
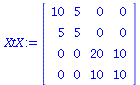 |
(18) |
| > |
XtY:=Multiply(Transpose(X),Y); |
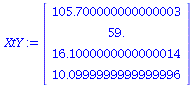 |
(19) |
| > |
beta:=LinearSolve(XtX,XtY); |
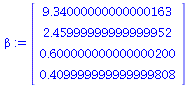 |
(20) |
Note that, contained here are the values for the two slopes we saw before(!)
The estimate for ![beta[3]](images/HypTestMultReg_37.gif)
Note this is (0,0,0,1) 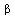 , so we take
, so we take 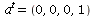 in our general formulas:
in our general formulas:
| > |
a:=Matrix([[0],[0],[0],[1]]): |
| > |
S2:=1/(10-(3+1))*(Multiply(Transpose(Y),Y)[1,1] - Multiply(Multiply(Transpose(beta),Transpose(X)),Y)[1,1]); |
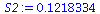 |
(21) |
| > |
t:=beta[4,1]/sqrt(S2*Multiply(Transpose(a),LinearSolve(XtX,a))[1,1]); |
 |
(22) |
With 10 - (3+1) = 6 d.f. this gives a p - value less than 0.05:
| > |
T6:=RandomVariable(StudentT(6)); |
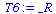 |
(23) |
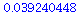 |
(24) |
So at 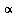 = .05 level, for instance, this would be sufficient evidence to reject
= .05 level, for instance, this would be sufficient evidence to reject
![H[0]](images/HypTestMultReg_45.gif) and conclude
and conclude ![`<>`(beta[3], 0)](images/HypTestMultReg_46.gif) . As above, this indicates that the rates of growth
. As above, this indicates that the rates of growth
of the two strains of bacteria are different.
![]() if the measurement is from type A, and
if the measurement is from type A, and ![]() if the measurement is from type B. Then
if the measurement is from type B. Then ![]() will indicate the time, and Y will
will indicate the time, and Y will ![]() +
+ ![]()
![]() (type A), we get
(type A), we get ![]()
![]() (type B), then we get
(type B), then we get ![]()
![]() =
= ![]() . We know
. We know ![]() :
: ![]() versus
versus ![]() :
: ![]() based on our general
based on our general 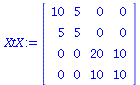
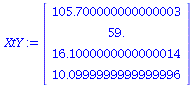
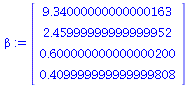
![]()
![]() , so we take
, so we take ![]() in our general formulas:
in our general formulas: ![]() = .05 level, for instance, this would be sufficient evidence to reject
= .05 level, for instance, this would be sufficient evidence to reject ![]() and conclude
and conclude ![]() . As above, this indicates that the rates of growth
. As above, this indicates that the rates of growth